Surface plasmon resonance (SPR) can be used to analyze all types of interactions ranging from protein-protein, protein-small molecule, protein-nucleic acid, protein-aptamer, protein-lipid, carbohydrate-protein and many more. One area of research is the study of aptamer binding kinetics, and many scientists are implementing techniques such as SPR to define the kinetic constants of these types of interactions.
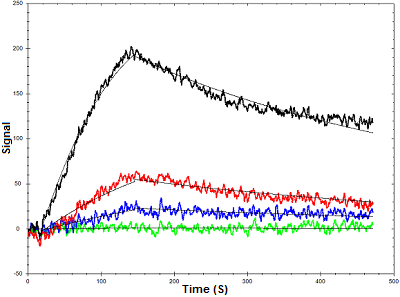
Figure 1. Binding curves of lysozyme binding to an aptamer using OpenSPR
So what is an aptamer and what makes them so special?
Jurassic Park taught us that DNA is key for bringing back the dinosaurs. But if you haven’t heard of aptamers yet, you’re in for a treat. This new area of study has a great deal of potential. Aptamers are a class of small nucleic acid molecules that are able to form secondary and tertiary structures capable of binding specifically to proteins and other cellular targets.You can view them as the antibody of the DNA world – because they function just like antibodies do.
But what makes aptamers so special? Well for starters, they are highly specific to their binding partner, non-immunogenic and relatively small in size. They are synthesized via high-throughput chemical synthesis and quality reveals very little batch-to-batch variation. These factors alone make aptamer production revolutionary in comparison to traditional antibody production techniques. Their high specificity and ease of modification for conjugation makes them ideal as targeting ligands and clinically relevant for diseases such as cancer, macular degeneration and HIV. Applications for aptamers are vast for basic science, as they could be used for purification procedures, biosensors, and as a replacement for traditional antibody techniques.
Aptamers naturally fold into unique structures with high specificity to their binding partner
Where do aptamers come from?
Aptamers are ligands that are identified from random libraries of DNA, RNA, or oligonucleotides by an in vitro selection process called “systematic evolution of ligands by exponential enrichment” (SELEX) technology. This technology is used to produce chemically modified oligonucleotides via a combinatorial chemistry technique in molecular biology. SELEX produces single-stranded DNA aptamers or RNA aptamers. SELEX aptamers have high target affinity for their ligand.
Why is it important to determine aptamer binding kinetics and why do researchers turn to SPR?
Since aptamers are meant to bind specifically to their ligand, it is important for researchers to understand the binding kinetics of the interaction. SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows you to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. Researchers use SPR for aptamer biosensor development, to test sequences specificity, quality control and validation, cell capture and to determine aptamer affinity. SPR provides quantitative data that researchers can use to fully characterize their aptamer interactions.
Example of an aptamer interaction analyzed using OpenSPR™
Below is an example of how OpenSPR™ was used to characterize the binding kinetics of an aptamer-lysozyme interaction. Lysozymes are enzymes that damage bacterial cell walls via glycoside hydrolysis. A lysozyme-specific aptamer was immobilized to a streptavidin sensor chip via a biotin tag and the binding kinetics were determined using four concentrations of lysozyme. The data was fit to a one-to-one binding diffusion corrected model. This data illustrates the value of surface plasmon resonance analysis for aptamer-protein interactions, as minimal samples were needed to extract powerful data. You can find the full study here.
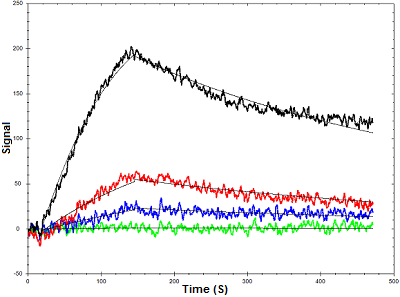
Figure 1. Binding curves of lysozyme binding to an aptamer using OpenSPR
